Identify trends and patterns in high-ranking molecular scaffolds using clustering algorithms in Spark™

Spark is a well-established software solution for computational bioisosteric replacement. In fact, our customers tell us it’s the best scaffold hopping tool they have ever used! Spark uses Cresset’s proprietary XED force field to assess similarity of molecular fragments to a known reference, allowing hundreds of new molecular designs to be generated in a matter of minutes. Production of large numbers of novel compounds poses a challenge to medicinal chemists, who must quickly assess and then gather insight from the data generated in this kind of experiment.
Clustering newly generated scaffolds in Spark allows you to group compounds based on chemotype. This allows patterns in the molecules to be easily identified, showing which chemistries best match the reference. With several options available, we briefly discuss here some of the key technical and practical considerations for compound clustering.
In the following example, Spark was used to find bioisosteres for the crystallographic ligand of PDB code, 1OIT, which is a cyclin-dependant kinase. Each newly generated molecule was assigned a score, which is displayed in the ‘Score’ column of the Clustered Results table.
The clustering algorithm can easily be adjusted via Project à Clustering Algorithm, then selecting Normal Clustering, Scaffold Clustering, Substituent Clustering, Scaffold-Substituent Clustering.
For each cluster, the highest scoring compound is defined as the ‘representative’. Where there are multiple compounds assigned to a cluster, the number of compounds listed in the ‘NC’ column will be greater than 1. The effect of changing the clustering algorithm in this can be seen clearly in Figure 1, as the number of compounds in each cluster changes significantly (circled in orange).
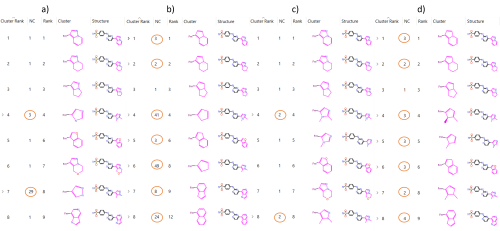
Figure 1. Spark Clustered Results Table group according to a) Normal Clustering, b) Scaffold Clustering, c) Substituent Clustering, d) Scaffold-Substituent Clustering.
Normal Clustering
The Normal Clustering method groups compounds in the same cluster if the path between connected atoms is the same, including all atoms in rings along the path. Different heteroatoms along the same 2D structure are different paths, meaning that (-CH2CH2-) is clustered differently to (-CH2NH-), despite the structural similarities.
Scaffold Clustering
In the Scaffold Clustering method, the same 2D structural approach to the Normal Clustering method is used, however all elements are converted to Carbon. This maintains the 2D structural classification present in the Normal Clustering method, however this method is insensitive to the presence of heteroatoms. This often generates larger clusters than the Normal Clustering method, as can be seen in Figure 1. Also shown in Figure 1 is the scaffold used to cluster the compounds. Comparison of the ‘Cluster’ column in Figures 1a and 1b shows that though the shape of the core scaffold remains the same between Normal and Scaffold Clustering, there are no heteroatoms present in the Scaffold Clustering. As an example of how a molecule may be differently clustered by Scaffold Clustering compared to Normal Clustering; the single molecule at Cluster Rank six in Figure 1a (Normal) is clustered as part of Cluster Rank two in Figure 1b (Scaffold).
Substituent Clustering
The Substituent Clustering method works similarly to Normal Clustering, however, this method is sensitive to ring substitutions. A benzyl replacement (-CH2-Ph) with para or meta substitution would therefore occupy the same cluster according in the Normal Clustering method, but different clusters according to the Substituent Clustering method. The Substituent Clustering method is sensitive to the substituent species, meaning that para (Ph-Cl) occupies a different cluster to para (Ph-Br). Comparison of Normal to Substituent Clustering from Figures 1a and 1c reports identical clustering for the first three compounds because there are no differences in ring-substituents. Cluster Rank four contains a different number of molecules when comparing Figures 1a and 1c, despite having the same representative compound. This is because the Normal method does not compare ring-substituents, which can be seen by visual comparison of the core scaffolds shown in the ‘Cluster’ column. In Substituent Clustering, the core scaffold contains methyl groups attached to the five-membered ring, which excludes one of the compounds assigned to this cluster by the Normal method.
Scaffold-Substituent Clustering
Finally, the Scaffold-Substituent Clustering method combines the prior two methods such that all elements are converted to Carbon before being clustered on ring substituent position, meaning sensitivity towards ring position, but not substituent species. Comparison of Cluster Rank four across Figure 1 demonstrates the differences between methods. Figure 1d shows the core scaffold used for the Scaffold-Substituent Clustering method. Figure 1d shows that although ring-substituents are included on the five-membered ring, there are no heteroatoms, which combines changes from Figures 1b and 1c, compared to Figure 1a.
Try Spark on your project
See the flexible licensing options for Spark and request an evaluation to generate highly innovative ideas in your project.

























